Understanding Dask’s meta keyword argument
By Pavithra Eswaramoorthy
If you have worked with Dask DataFrames or Dask Arrays, you have probably come across the meta keyword argument. Perhaps, while using methods like apply():
import dask
import pandas as pd
ddf = dask.datasets.timeseries()
def my_custom_arithmetic(r):
if r.id > 1000:
return r.x * r.x + r.y + r.y
else:
return 0
ddf["my_computation"] = ddf.apply(
my_custom_arithmetic, axis=1, meta=pd.Series(dtype="float64")
)
ddf.head()
# Output:
#
# id name x y my_computation
# timestamp
# 2000-01-01 00:00:00 1055 Victor -0.575374 0.868320 2.067696
# 2000-01-01 00:00:01 994 Zelda 0.963684 0.972240 0.000000
# 2000-01-01 00:00:02 982 George -0.997531 -0.876222 0.000000
# 2000-01-01 00:00:03 981 Ingrid 0.852159 -0.419733 0.000000
# 2000-01-01 00:00:04 1029 Jerry -0.839431 -0.736572 -0.768500
You might have also seen one or more of the following warnings/errors:
UserWarning: You did not provide metadata, so Dask is running your function on a small dataset to guess output types. It is possible that Dask will guess incorrectly.
UserWarning: `meta` is not specified, inferred from partial data. Please provide `meta` if the result is unexpected.
ValueError: Metadata inference failed in …
If the above messages look familiar, this blog post is for you. :)
We will discuss:
- what the is
metakeyword argument, - why does Dask need
meta, and - how to use it effectively.
We will look at meta mainly in the context of Dask DataFrames, however, similar principles also apply to Dask Arrays.
What is meta?
Before answering this, let’s quickly discuss Dask DataFrames.
A Dask DataFrame is a lazy object composed of multiple pandas DataFrames, where each pandas DataFrame is called a “partition”. These are stacked along the index and Dask keeps track of these partitions using “divisions”, which is a tuple representing the start and end index of each partition.
When you create a Dask DataFrame, you usually see something like the following:
>>> import pandas as pd
>>> import dask.dataframe as dd
>>> ddf = dd.DataFrame.from_dict(
... {
... "x": range(6),
... "y": range(10, 16),
... },
... npartitions=2,
... )
>>> ddf
Dask DataFrame Structure:
x y
npartitions=2
0 int64 int64
3 ... ...
5 ... ...
Dask Name: from_pandas, 2 tasks
Here, Dask has created the structure of the DataFrame using some “metadata” information about the column names and their datatypes. This metadata information is called meta. Dask uses meta for understanding Dask operations and creating accurate task graphs (i.e., the logic of your computation).
The meta keyword argument in various Dask DataFrame functions allows you to explicitly share this metadata information with Dask. Note that the keyword argument is concerned with the metadata of the output of those functions.
Why does Dask need meta?
Dask computations are evaluated lazily. This means Dask creates the logic and flow, called task graph, of the computation immediately, but evaluates them only when necessary – usually, on calling .compute().
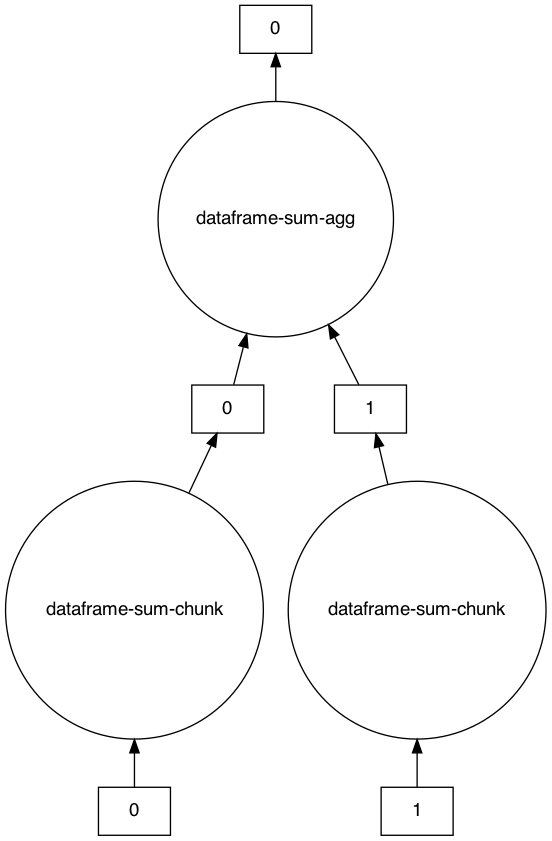
An example task graph generated to compute the sum of the DataFrame:
>>> s = ddf.sum()
>>> s
Dask Series Structure:
npartitions=1
x int64
y ...
dtype: int64
Dask Name: dataframe-sum-agg, 5 tasks
>>> s.visualize()
>>> s.compute()
x 15
y 75
dtype: int64
This is a single operation, but Dask workflows usually have multiple such operation chained together. Therefore, to create the task graph effectively, Dask needs to know the strucutre and datatypes of the DataFame after each operation. Especially because Dask does not know the actual values/structure of the DataFrame yet.
This is where meta is comes in.
In the above example, the Dask DataFrame changed into a Series after sum(). Dask knows this (even before we call compute()) only becasue of meta.
Internally, meta is represented as an empty pandas DataFrame or Series, which has the same structure as the Dask DataFrame. To learn more about how meta is defined internally, check out the DataFrame Internal Design documentation.
To see the actual metadata information for a collection, you can look at the ._meta attribute[1]:
>>> s._meta
Series([], dtype: int64)
How to specify meta?
You can specify meta in a few different ways, but the recommended way for Dask DataFrame is:
“An empty
pd.DataFrameorpd.Seriesthat matches the dtypes and column names of the output.”
For example:
>>> meta_df = pd.DataFrame(columns=["x", "y"], dtype=int)
>>> meta_df
Empty DataFrame
Columns: [x, y]
Index: []
>>> ddf2 = ddf.apply(lambda x: x, axis=1, meta=meta_df).compute()
>>> ddf2
x y
0 0 0
1 1 1
2 2 2
3 0 3
4 1 4
5 2 5
The other ways you can describe meta are:
-
For a DataFrame, you can specify
metaas a:- Python dictionary:
{column_name_1: dtype_1, column_name_2: dtype_2, …} - Iterable of tuples:
[(column_name_1, dtype_1), (columns_name_2, dtype_2, …)]
Note that while describing
metaas shown above, using a dictionary or iterable of tuples, the order in which you mention column names is important. Dask will use the same order to create the pandas DataFrame formeta. If the orders don’t match, you will see the following error:ValueError: The columns in the computed data do not match the columns in the provided metadata - Python dictionary:
-
For a Series output, you can specify
metausing a single tuple:(coulmn_name, dtype)
You should not describe meta using just a dtype (like: meta="int64"), even for scalar outputs. If you do, you will see the following warning:
FutureWarning: Meta is not valid, `map_partitions` and `map_overlap` expects output to be a pandas object. Try passing a pandas object as meta or a dict or tuple representing the (name, dtype) of the columns. In the future the meta you passed will not work.
During operations like map_partitions or apply (which uses map_partitions internally), Dask coerces the scalar output of each partition into a pandas object. So, the output of functions that take meta will never be scalar.
For example:
>>> ddf = ddf.repartition(npartitions=1)
>>> result = ddf.map_partitions(lambda x: len(x)).compute()
>>> type(result)
pandas.core.series.Series
Here, the Dask DataFrame ddf has only one partition. Hence, len(x) on that one partition would result in a scalar output of integer dtype. However, when we compute it, we see a pandas Series. This confirms that Dask is coercing the outputs to pandas objects.
Another note, Dask Arrays may not always do this conversion. You can look at the API reference for your particular Array operation for details.
>>> import numpy as np
>>> import dask.array as da
>>> my_arr = da.random.random(10)
>>> my_arr.map_blocks(lambda x: len(x)).compute()
10
meta does not force the structure or dtypes
meta can be thought of as a suggestion to Dask. Dask uses this meta to generate the task graph until it can infer the actual metadata from the values. It does not force the output to have the structure or dtype of the specified meta.
Consider the following example, and remember that we defined ddf with x and y column names in the previous sections.
If we provide different column names (a and b) in the meta description, Dask uses these new names to create the task graph:
>>> meta_df = pd.DataFrame(columns=["a", "b"], dtype=int)
>>> result = ddf.apply(lambda x:x*x, axis=1, meta=meta_df)
>>> result
Dask DataFrame Structure:
a b
npartitions=2
0 int64 int64
3 ... ...
5 ... ...
Dask Name: apply, 4 tasks
However, if we compute result, we will get the following error:
>>> result.compute()
ValueError: The columns in the computed data do not match the columns in the provided metadata
Extra: ['x', 'y']
Missing: ['a', 'b']
While computing, Dask evaluates the actual metadata with columns x and y. This does not match the meta that we provided, and hence, Dask raises a helpful error message. Notice how Dask does not change the output to have a and b here, rather uses a and b column names only for intermediate task graphs.
Using ._meta directly
In some rare case, you can also set the ._meta attribute[1] directly for a Dask DataFrame. For example, if the DataFrame was created with incorrect dtypes, like:
>>> ddf = dd.DataFrame.from_dict(
... {
... "x": range(6),
... "y": range(10, 16),
... },
... dtype="object", # Note the “object” dtype
... npartitions=2,
... )
>>> ddf
Dask DataFrame Structure:
x y
npartitions=2
0 object object
3 ... ...
5 ... ...
Dask Name: from_pandas, 2 tasks
The values are clearly integers but the dtype says object, so we can’t perform integer operations like addition:
>>> add = ddf + 2
ValueError: Metadata inference failed in `add`.
Original error is below:
------------------------
TypeError('can only concatenate str (not "int") to str')
Here, we can explicitly define ._meta[1]:
>>> ddf._meta = pd.DataFrame(columns=["x", "y"], dtype="int64")
Then, perform the addition:
>>> add = ddf + 2
>>> add
Dask DataFrame Structure:
x y
npartitions=2
0 int64 int64
3 ... ...
5 ... ...
Dask Name: add, 4 tasks
Thanks for reading!
Have you run into issues with meta before? Please let us know on Discourse, and we will consider including it here, or updating the Dask documentation. :)
[1] NOTE: ._meta is not a public property, so we recommend using it only when necessary. There is an ongoing discussion around creating public methods to get, set, and view the information in ._meta, and this blog post will be updated to use the public methods when they’re created.
blog comments powered by Disqus