Mosaic Image Fusion
By Volker Hilsenstein, Marvin Albert, and Genevieve Buckley
Executive Summary
This blogpost shows a case study where a researcher uses Dask for mosaic image fusion.
Mosaic image fusion is when you combine multiple smaller images taken at known locations and stitch them together into a single image with a very large field of view. Full code examples are available on GitHub from the DaskFusion repository:
https://github.com/VolkerH/DaskFusion
The problem
Image mosaicing in microscopy
In optical microscopy, a single field of view captured with a 20x objective typically has a diagonal on the order of a few 100 μm (exact dimensions depend on other parts of the optical system, including the size of the camera chip). A typical sample slide has a size of 25mm by 75mm. Therefore, when imaging a whole slide, one has to acquire hundreds of images, typically with some overlap between individual tiles. With increasing magnification, the required number of images increases accordingly.
To obtain an overview one has to fuse this large number of individual image tiles into a large mosaic image. Here, we assume that the information required for positioning and alignment of the individual image tiles is known. In the example presented here, this information is available as metadata recorded by the microscope, namely the microscope stage position and the pixel scale. Alternatively, this information could also be derived from the image data directly, e.g. through a registration step that matches corresponding image features in the areas where tiles overlap.
The solution
The array that can hold the resulting mosaic image will often have a size that is too large
to fit in RAM, therefore we will use Dask arrays and the map_blocks function to enable
out-of-core processing. The map_blocks
function will process smaller blocks (a.k.a chunks) of the output array individually, thus eliminating the need to
hold the whole output array in memory. If sufficient resources are available, dask will also distribute the processing of blocks across several workers,
thus we also get parallel processing for free, which can help speed up the fusion process.
Typically whenever we want to join Dask arrays, we use Stack, Concatenate, and Block. However, these are not good tools for mosaic image fusion, because:
- The image tiles will be be overlapping,
- Tiles may not be positioned on an exact grid and will typically also have slight rotations as the alignment of stage and camera is not perfect. In the most general case, for example in panaromic photo mosaics, individual image tiles could be arbitrarily rotated or skewed.
The starting point for this mosaic prototype was some code that reads in the stage metadate for all tiles and calculates an affine transformation for each tile that would place it at the correct location in the output array.
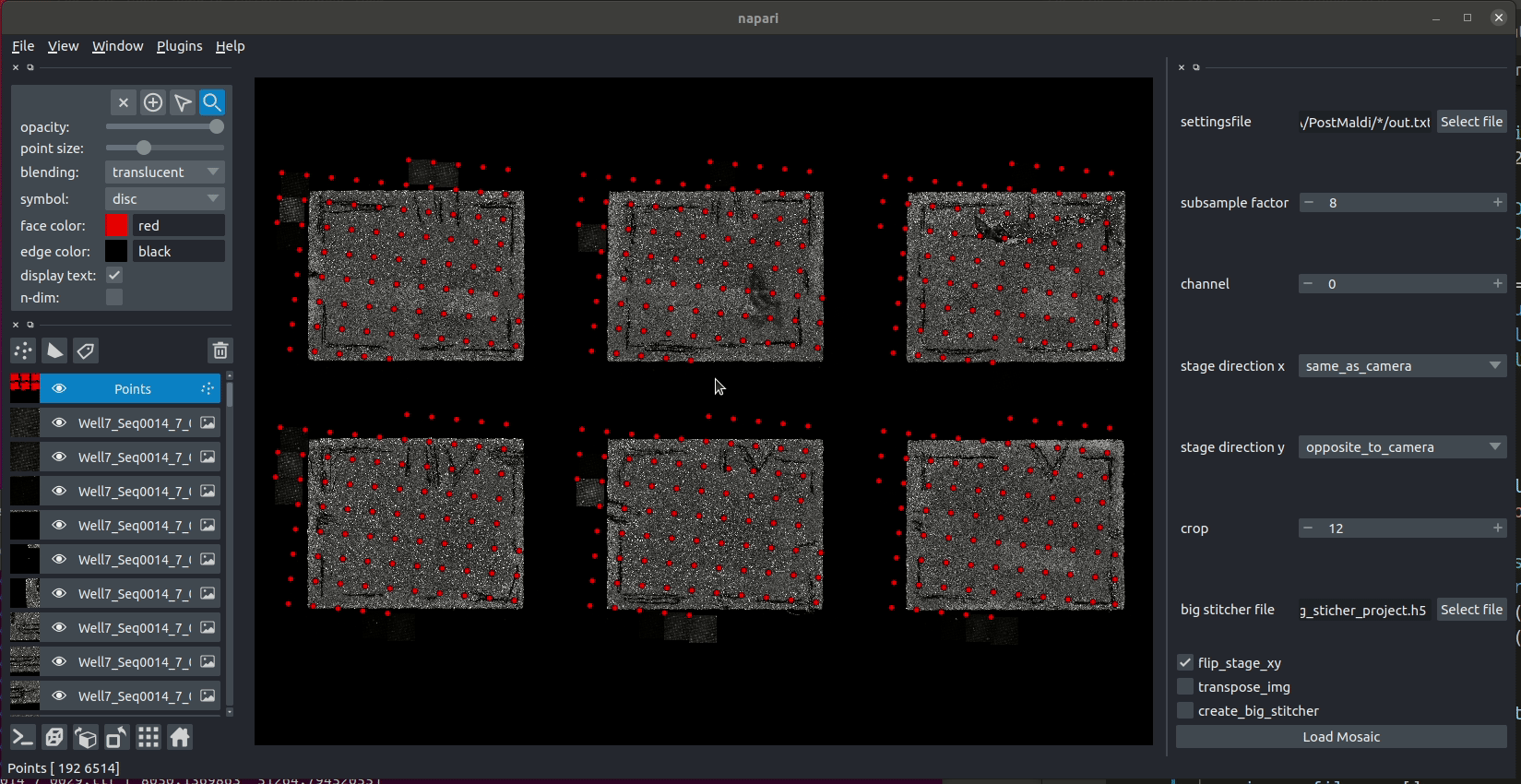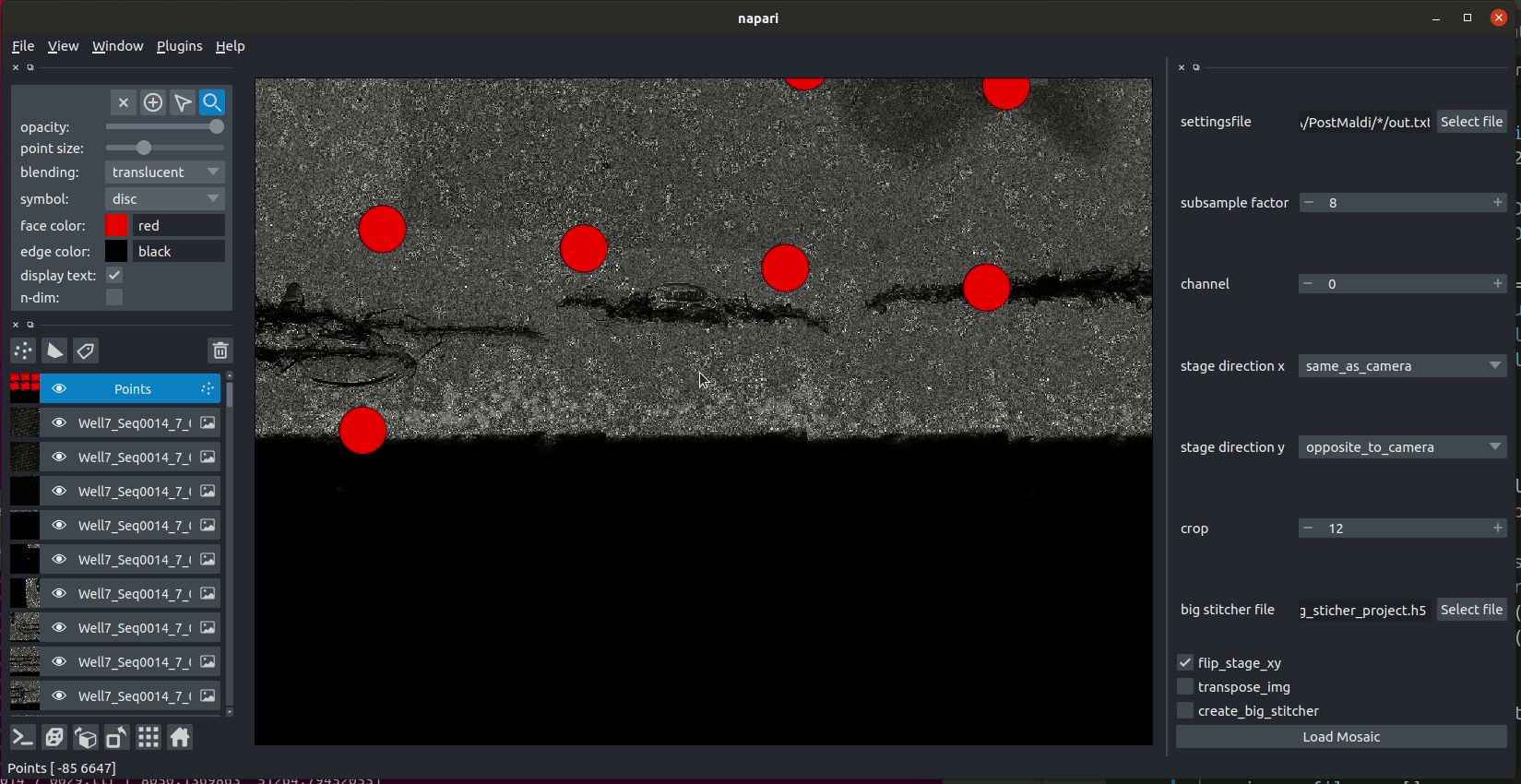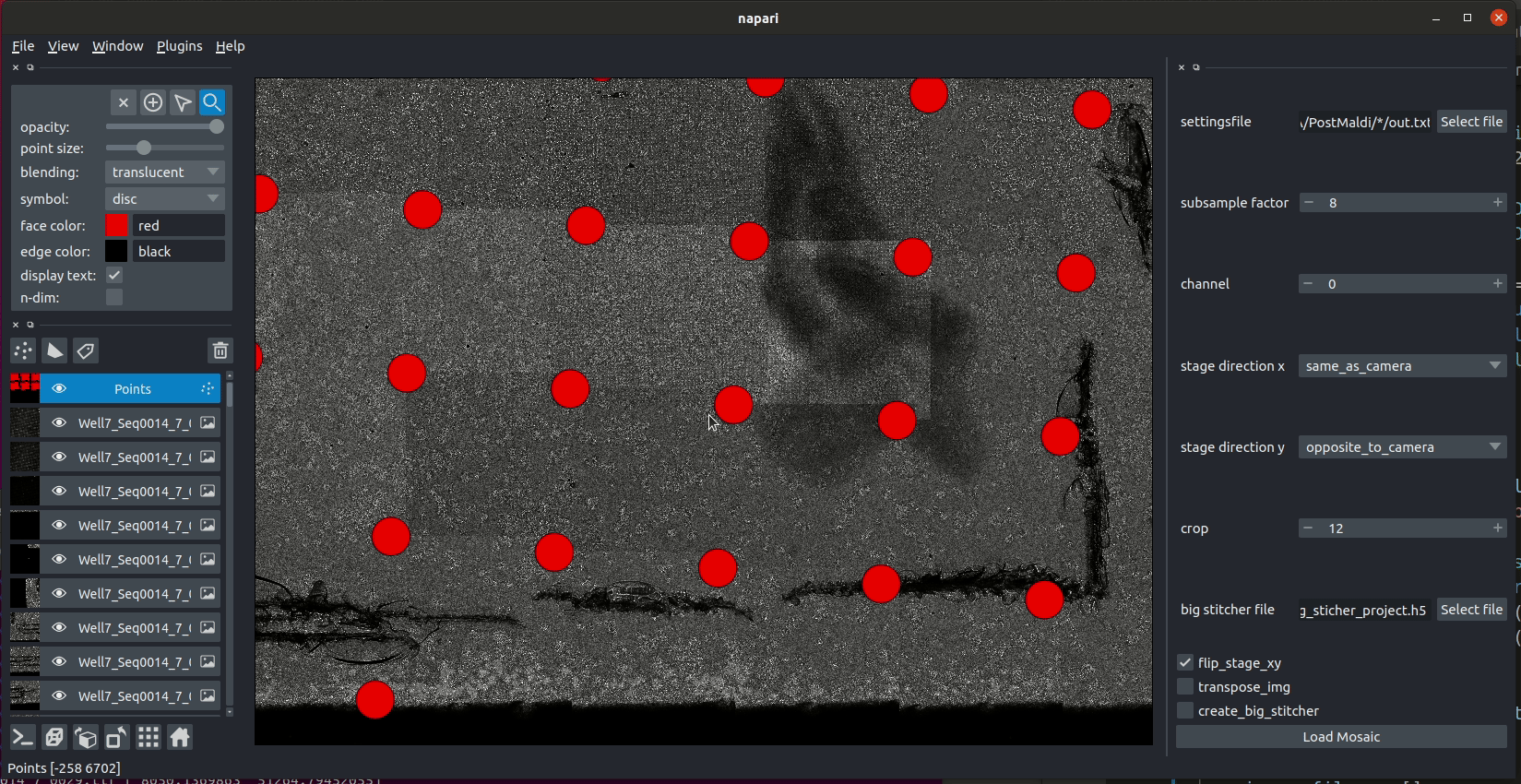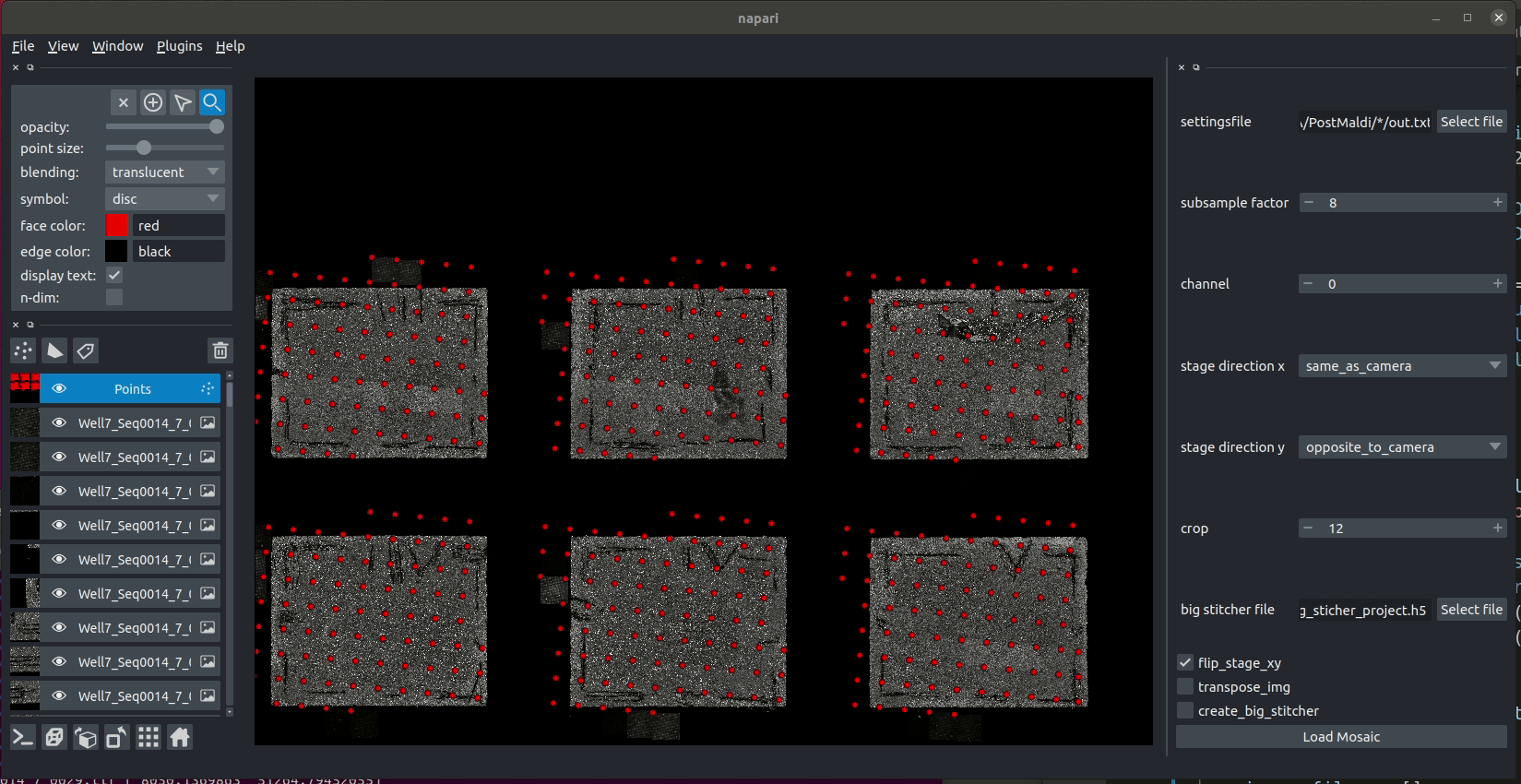
The image below shows preliminary work placing mosaic image tiles into the correct positions using the napari image viewer. Shown here is a small example with 63 image tiles.
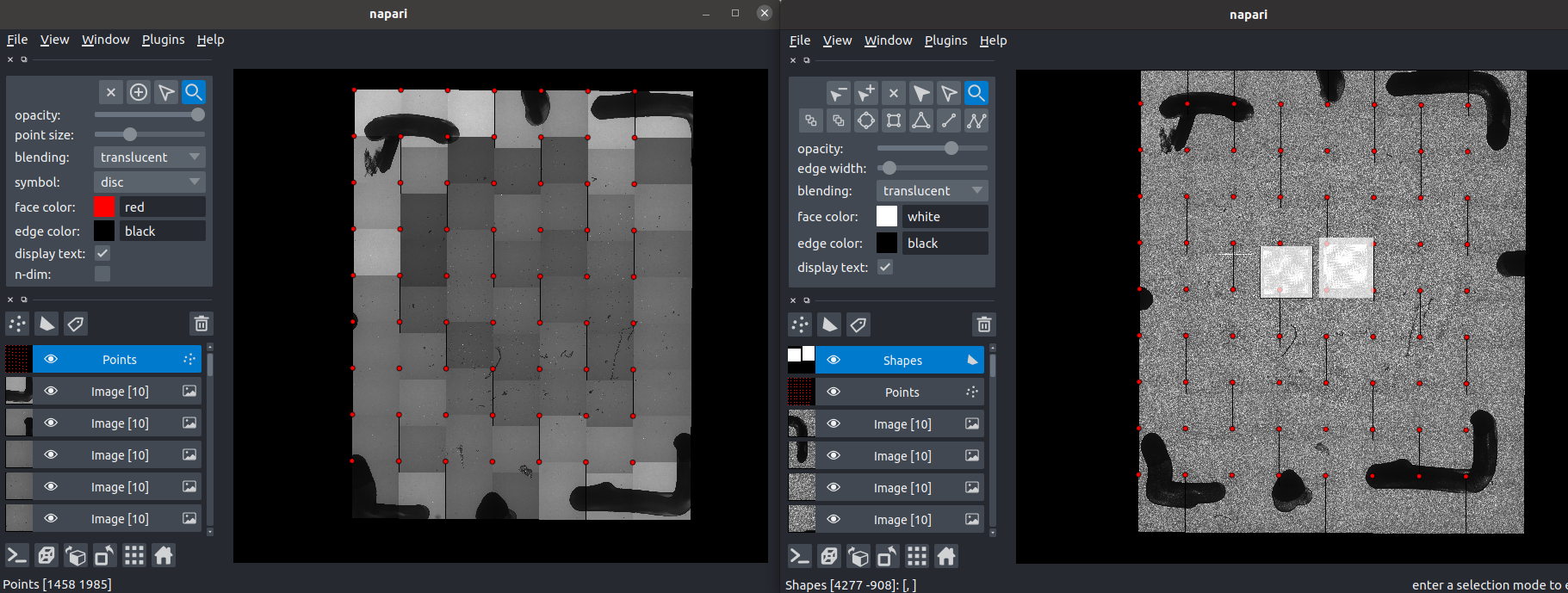
And here is an animation of placing the individual tiles.
To leverage processing with Dask we created a fuse function that generates a small block of the final mosaic and is invoked by map_blocks for each chunk of the output array.
On each invocation of the fuse function map_blocks passes a dictionary (block_info). From the Dask documentation:
Your block function gets information about where it is in the array by accepting a special
block_infoorblock_idkeyword argument.
The basic outline of the fuse function of the mosaic workflow is as follows.
For each chunk of the output array:
- Determine which source image tiles intersect with the chunk.
- Adjust the image tiles’ affine transformations to take the offset of the chunk within the array into account.
- Load all intersectiong image tiles and apply their respective adjusted affine transformation to map them into the chunk.
- Blend the tiles using a simple maximum projection.
- Return the blended chunk.
Using a maximum projection to blend areas with overlapping tiles can lead to artifacts such as ghost images and visible tile seams, so you would typically want to use something more sophisticated in production.
Results
For datasets with many image tiles (~500-1000 tiles), we could speed up the mosaic generation from several hours to tens of minutes using this Dask based method (compared to a previous workflow using ImageJ plugins runnning on the same workstation). Due to Dask’s ability to handle data out-of-core and chunked array storage using zarr it is also possible to run the fusion on hardware with limited RAM.
Finally, we have the final mosaic fusion result.
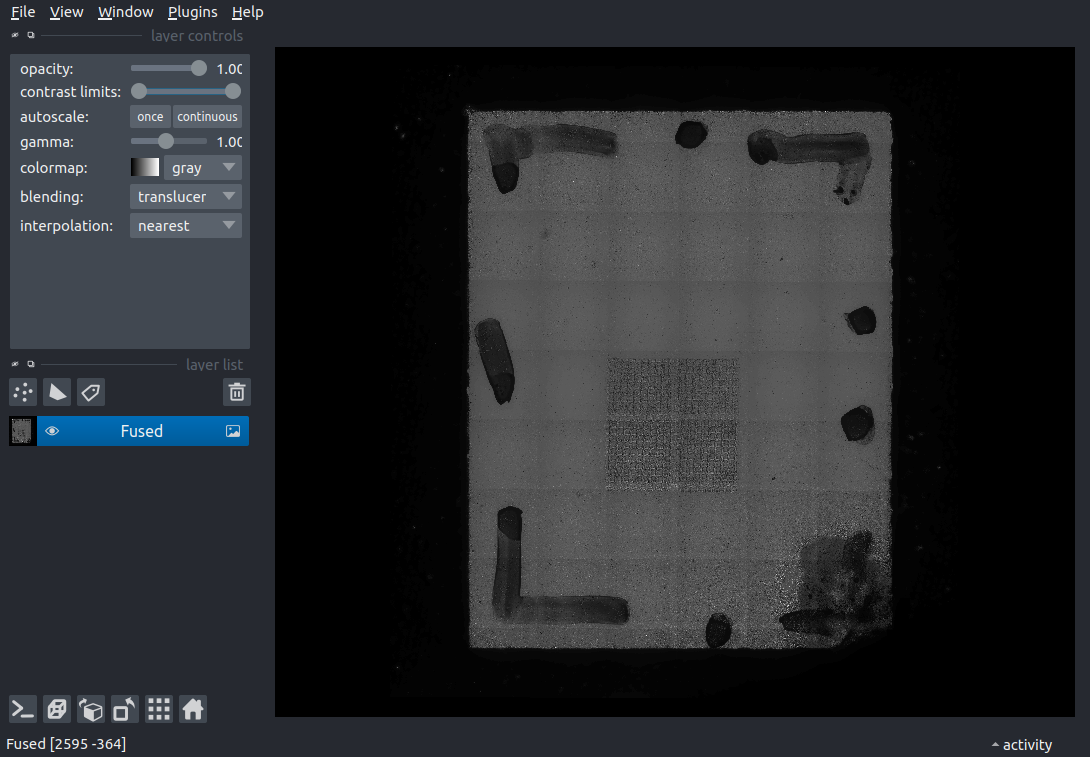
Code
Code relatiing to this mosaic image fusion project can be found in the DaskFusion GitHub repository here:
https://github.com/VolkerH/DaskFusion
There is a self-contained example available in this notebook, which downloads reduced-size example data to demonstrate the process.
What’s next?
Currently, the DaskFusion code is a proof of concept for single-channel 2D images and simple maximum projection for blending the tiles in overlapping areas, it is not production code. However, the same principle can be used for fusing multi-channel image volumes, such as from Light-Sheet data if the tile chunk intersection calculation is extended to higher-dimensional arrays. Such even larger datasets will benefit even more from leveraging dask, as the processing can be distributed across multiple nodes of a HPC cluster using dask jobqueue.
Also see
Marvin’s lightning talk on multi-view image fusion: 15 minute video available here on YouTube
The GitHub repository MVRegFus that Marvin talks about in the video is available here: https://github.com/m-albert/MVRegFus
The napari-lazy-openslide visualization plugin by Trevor Manz: “An experimental plugin to lazily load multiscale whole-slide tiff images with openslide and dask.”
For further information on alternative approaches to image stitching:
- ASHLAR: Alignment by Simultaneous Harmonization of Layer / Adjacency Registration
- Microscopy Image Stitching Tool (MIST)
- The m2stitch python package by Yohsuke T. Fukai: “Provides robust stitching of tiled microscope images on a regular grid” (based on the MIST algorithm)
Acknowledgements
This computational work was done by Volker Hilsenstein, in conjunction with Marvin Albert. Volker Hilsenstein is a scientific software developer at EMBL in Theodore Alexandrov’s lab with a focus on spatial metabolomics and bio-image analysis.
The sample images were prepared and imaged by Mohammed Shahraz from the Alexandrov lab at EMBL Heidelberg.
Genevieve Buckley and Volker Hilsenstein wrote this blogpost.
blog comments powered by Disqus