Comparing Dask-ML and Ray Tune's Model Selection Algorithms Modern hyperparameter optimizations, Scikit-Learn support, framework support and scaling to many machines.
By Scott Sievert (University of Wisconsin–Madison)
Hyperparameter optimization is the process of deducing model parameters that can’t be learned from data. This process is often time- and resource-consuming, especially in the context of deep learning. A good description of this process can be found at “Tuning the hyper-parameters of an estimator,” and the issues that arise are concisely summarized in Dask-ML’s documentation of “Hyper Parameter Searches.”
There’s a host of libraries and frameworks out there to address this problem. Scikit-Learn’s module has been mirrored in Dask-ML and auto-sklearn, both of which offer advanced hyperparameter optimization techniques. Other implementations that don’t follow the Scikit-Learn interface include Ray Tune, AutoML and Optuna.
Ray recently provided a wrapper to Ray Tune that mirrors the Scikit-Learn API called tune-sklearn (docs, source). The introduction of this library states the following:
Cutting edge hyperparameter tuning techniques (Bayesian optimization, early stopping, distributed execution) can provide significant speedups over grid search and random search.
However, the machine learning ecosystem is missing a solution that provides users with the ability to leverage these new algorithms while allowing users to stay within the Scikit-Learn API. In this blog post, we introduce tune-sklearn [Ray’s tuning library] to bridge this gap. Tune-sklearn is a drop-in replacement for Scikit-Learn’s model selection module with state-of-the-art optimization features.
This claim is inaccurate: for over a year Dask-ML has provided access to “cutting edge hyperparameter tuning techniques” with a Scikit-Learn compatible API. To correct their statement, let’s look at each of the features that Ray’s tune-sklearn provides, and compare them to Dask-ML:
Here’s what [Ray’s] tune-sklearn has to offer:
- Consistency with Scikit-Learn API …
- Modern hyperparameter tuning techniques …
- Framework support …
- Scale up … [to] multiple cores and even multiple machines.
[Ray’s] Tune-sklearn is also fast.
Dask-ML’s model selection module has every one of the features:
- Consistency with Scikit-Learn API: Dask-ML’s model selection API mirrors the Scikit-Learn model selection API.
- Modern hyperparameter tuning techniques: Dask-ML offers state-of-the-art hyperparameter tuning techniques.
- Framework support: Dask-ML model selection supports many libraries including Scikit-Learn, PyTorch, Keras, LightGBM and XGBoost.
- Scale up: Dask-ML supports distributed tuning (how could it not?) and larger-than-memory datasets.
Dask-ML is also fast. In “Speed” we show a benchmark between Dask-ML, Ray and Scikit-Learn:
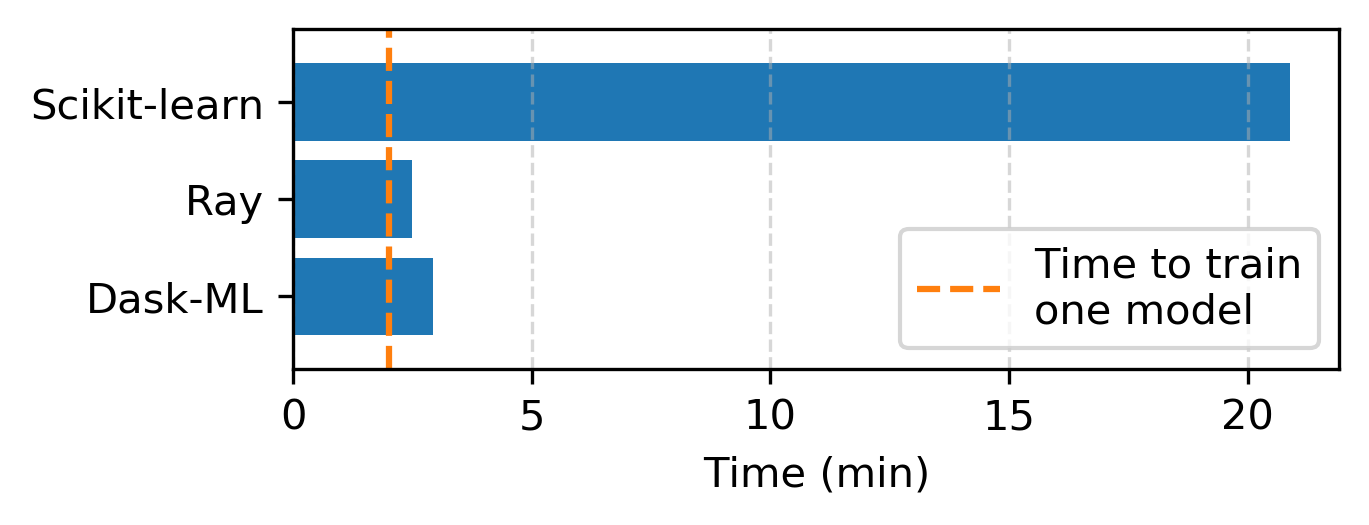
Only time-to-solution is relevant; all of these methods produce similar model scores. See “Speed” for details.
Now, let’s walk through the details on how to use Dask-ML to obtain the 5 features above.
Consistency with the Scikit-Learn API
Dask-ML is consistent with the Scikit-Learn API.
Here’s how to use Scikit-Learn’s, Dask-ML’s and Ray’s tune-sklearn hyperparameter optimization:
## Trimmed example; see appendix for more detail
from sklearn.model_selection import RandomizedSearchCV
search = RandomizedSearchCV(model, params, ...)
search.fit(X, y)
from dask_ml.model_selection import HyperbandSearchCV
search = HyperbandSearchCV(model, params, ...)
search.fit(X, y, classes=[0, 1])
from tune_sklearn import TuneSearchCV
search = TuneSearchCV(model, params, ...)
search.fit(X, y, classes=[0, 1])
The definitions of model and params follow the normal Scikit-Learn
definitions as detailed in the appendix.
Clearly, both Dask-ML and Ray’s tune-sklearn are Scikit-Learn compatible. Now let’s focus on how each search performs and how it’s configured.
Modern hyperparameter tuning techniques
Dask-ML offers state-of-the-art hyperparameter tuning techniques in a Scikit-Learn interface.
The introduction of Ray’s tune-sklearn made this claim:
tune-sklearn is the only Scikit-Learn interface that allows you to easily leverage Bayesian Optimization, HyperBand and other optimization techniques by simply toggling a few parameters.
The state-of-the-art in hyperparameter optimization is currently
“Hyperband.” Hyperband reduces the amount of computation
required with a principled early stopping scheme; past that, it’s the same as
Scikit-Learn’s popular RandomizedSearchCV.
Hyperband works. As such, it’s very popular. After the introduction of Hyperband in 2016 by Li et. al, the paper has been cited over 470 times and has been implemented in many different libraries including Dask-ML, Ray Tune, keras-tune, Optuna, AutoML,1 and Microsoft’s NNI. The original paper shows a rather drastic improvement over all the relevant implementations,2 and this drastic improvement persists in follow-up works.3 Some illustrative results from Hyperband are below:
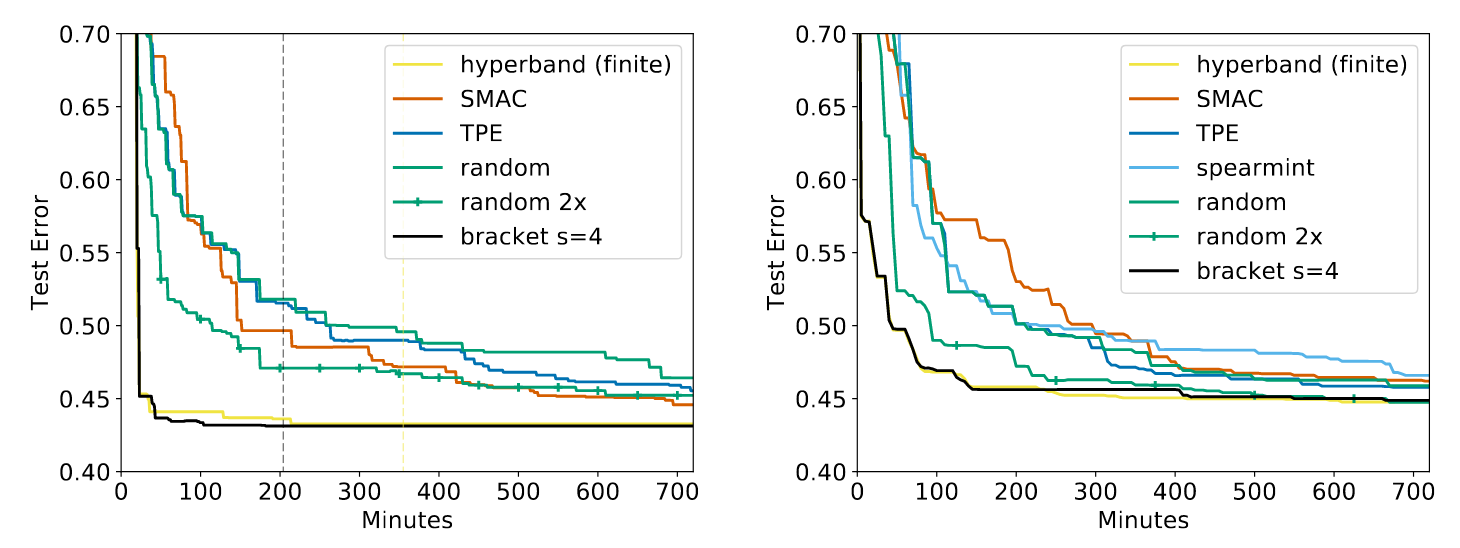
All algorithms are configured to do the same amount of work except “random 2x” which does twice as much work. “hyperband (finite)” is similar Dask-ML’s default implementation, and “bracket s=4” is similar to Ray’s default implementation. “random” is a random search. SMAC,4 spearmint,5 and TPE6 are popular Bayesian algorithms.
Hyperband is undoubtedly a “cutting edge” hyperparameter optimization technique. Dask-ML and Ray offer Scikit-Learn implementations of this algorithm that rely on similar implementations, and Dask-ML’s implementation also has a rule of thumb for configuration. Both Dask-ML’s and Ray’s documentation encourages use of Hyperband.
Ray does support using their Hyperband implementation on top of a technique called Bayesian sampling. This changes the hyperparameter sampling scheme for model initialization. This can be used in conjunction with Hyperband’s early stopping scheme. Adding this option to Dask-ML’s Hyperband implementation is future work for Dask-ML.
Framework support
Dask-ML model selection supports many libraries including Scikit-Learn, PyTorch, Keras, LightGBM and XGBoost.
Ray’s tune-sklearn supports these frameworks:
tune-sklearn is used primarily for tuning Scikit-Learn models, but it also supports and provides examples for many other frameworks with Scikit-Learn wrappers such as Skorch (Pytorch), KerasClassifiers (Keras), and XGBoostClassifiers (XGBoost).
Clearly, both Dask-ML and Ray support the many of the same libraries.
However, both Dask-ML and Ray have some qualifications. Certain libraries don’t
offer an implementation of partial_fit,7 so not all of the modern
hyperparameter optimization techniques can be offered. Here’s a table comparing
different libraries and their support in Dask-ML’s model selection and Ray’s
tune-sklearn:
| Model Library | Dask-ML support | Ray support | Dask-ML: early stopping? | Ray: early stopping? |
|---|---|---|---|---|
| Scikit-Learn | ✔ | ✔ | ✔* | ✔* |
| PyTorch (via Skorch) | ✔ | ✔ | ✔ | ✔ |
| Keras (via SciKeras) | ✔ | ✔ | ✔** | ✔** |
| LightGBM | ✔ | ✔ | ❌ | ❌ |
| XGBoost | ✔ | ✔ | ❌ | ❌ |
* Only for the models that implement partial_fit.
** Thanks to work by the Dask developers around scikeras#24.
By this measure, Dask-ML and Ray model selection have the same level of
framework support. Of course, Dask has tangential integration with LightGBM and
XGBoost through Dask-ML’s xgboost module and dask-lightgbm.
Scale up
Dask-ML supports distributed tuning (how could it not?), aka parallelization across multiple machines/cores. In addition, it also supports larger-than-memory data.
[Ray’s] Tune-sklearn leverages Ray Tune, a library for distributed hyperparameter tuning, to efficiently and transparently parallelize cross validation on multiple cores and even multiple machines.
Naturally, Dask-ML also scales to multiple cores/machines because it relies on Dask. Dask has wide support for different deployment options that span from your personal machine to supercomputers. Dask will very likely work on top of any computing system you have available, including Kubernetes, SLURM, YARN and Hadoop clusters as well as your personal machine.
Dask-ML’s model selection also scales to larger-than-memory datasets, and is thoroughly tested. Support for larger-than-memory data is untested in Ray, and there are no examples detailing how to use Ray Tune with the distributed dataset implementations in PyTorch/Keras.
In addition, I have benchmarked Dask-ML’s model selection module to see how the
time-to-solution is affected by the number of Dask workers in “Better and
faster hyperparameter optimization with Dask.” That is, how does the
time to reach a particular accuracy scale with the number of workers $P$? At
first, it’ll scale like $1/P$ but with large number of workers the serial
portion will dictate time to solution according to Amdahl’s Law. Briefly, I
found Dask-ML’s HyperbandSearchCV speedup started to saturate around 24
workers for a particular search.
Speed
Both Dask-ML and Ray are much faster than Scikit-Learn.
Ray’s tune-sklearn runs some benchmarks in the introduction with the
GridSearchCV class found in Scikit-Learn and Dask-ML. A more fair benchmark
would be use Dask-ML’s HyperbandSearchCV because it is almost the same as the
algorithm in Ray’s tune-sklearn. To be specific, I’m interested in comparing
these methods:
- Scikit-Learn’s
RandomizedSearchCV. This is a popular implementation, one that I’ve bootstrapped myself with a custom model. - Dask-ML’s
HyperbandSearchCV. This is an early stopping technique forRandomizedSearchCV. - Ray tune-sklearn’s
TuneSearchCV. This is a slightly different early stopping technique thanHyperbandSearchCV’s.
Each search is configured to perform the same task: sample 100 parameters and
train for no longer than 100 “epochs” or passes through the
data.8 Each estimator is configured as their respective
documentation suggests. Each search uses 8 workers with a single cross
validation split, and a partial_fit call takes one second with 50,000
examples. The complete setup can be found in the appendix.
Here’s how long each library takes to complete the same search:

Notably, we didn’t improve the Dask-ML codebase for this benchmark, and ran the code as it’s been for the last year.9 Regardless, it’s possible that other artifacts from biased benchmarks crept into this benchmark.
Clearly, Ray and Dask-ML offer similar performance for 8 workers when compared with Scikit-Learn. To Ray’s credit, their implementation is ~15% faster than Dask-ML’s with 8 workers. We suspect that this performance boost comes from the fact that Ray implements an asynchronous variant of Hyperband. We should investigate this difference between Dask and Ray, and how each balances the tradeoffs, number FLOPs vs. time-to-solution. This will vary with the number of workers: the asynchronous variant of Hyperband provides no benefit if used with a single worker.
Dask-ML reaches scores quickly in serial environments, or when the number of workers is small. Dask-ML prioritizes fitting high scoring models: if there are 100 models to fit and only 4 workers available, Dask-ML selects the models that have the highest score. This is most relevant in serial environments;10 see “Better and faster hyperparameter optimization with Dask” for benchmarks. This feature is omitted from this benchmark, which only focuses on time to solution.
Conclusion
Dask-ML and Ray offer the same features for model selection: state-of-the-art features with a Scikit-Learn compatible API, and both implementations have fairly wide support for different frameworks and rely on backends that can scale to many machines.
In addition, the Ray implementation has provided motivation for further development, specifically on the following items:
- Adding support for more libraries, including Keras (dask-ml#696, dask-ml#713, scikeras#24). SciKeras is a Scikit-Learn wrapper for Keras that (now) works with Dask-ML model selection because SciKeras models implement the Scikit-Learn model API.
- Better documenting the models that Dask-ML supports (dask-ml#699). Dask-ML supports any model that implement the Scikit-Learn interface, and there are wrappers for Keras, PyTorch, LightGBM and XGBoost. Now, Dask-ML’s documentation prominently highlights this fact.
The Ray implementation has also helped motivate and clarify future work. Dask-ML should include the following implementations:
- A Bayesian sampling scheme for the Hyperband implementation that’s similar to Ray’s and BOHB’s (dask-ml#697).
- A configuration of
HyperbandSearchCVthat’s well-suited for exploratory hyperparameter searches. An initial implementation is in dask-ml#532, which should be benchmarked against Ray.
Luckily, all of these pieces of development are straightforward modifications because the Dask-ML model selection framework is pretty flexible.
Thank you Tom Augspurger, Matthew Rocklin, Julia Signell, and Benjamin Zaitlen for your feedback, suggestions and edits.
Appendix
Benchmark setup
This is the complete setup for the benchmark between Dask-ML, Scikit-Learn and Ray. Complete details can be found at stsievert/dask-hyperband-comparison.
Let’s create a dummy model that takes 1 second for a partial_fit call with
50,000 examples. This is appropriate for this benchmark; we’re only interested
in the time required to finish the search, not how well the models do.
Scikit-learn, Ray and Dask-ML have have very similar methods of choosing
hyperparameters to evaluate; they differ in their early stopping techniques.
from scipy.stats import uniform
from sklearn.model_selection import make_classification
from benchmark import ConstantFunction # custom module
# This model sleeps for `latency * len(X)` seconds before
# reporting a score of `value`.
model = ConstantFunction(latency=1 / 50e3, max_iter=max_iter)
params = {"value": uniform(0, 1)}
# This dummy dataset mirrors the MNIST dataset
X_train, y_train = make_classification(n_samples=int(60e3), n_features=784)
This model will take 2 minutes to train for 100 epochs (aka passes through the data). Details can be found at stsievert/dask-hyperband-comparison.
Let’s configure our searches to use 8 workers with a single cross-validation split:
from sklearn.model_selection import RandomizedSearchCV, ShuffleSplit
split = ShuffleSplit(test_size=0.2, n_splits=1)
kwargs = dict(cv=split, refit=False)
search = RandomizedSearchCV(model, params, n_jobs=8, n_iter=n_params, **kwargs)
search.fit(X_train, y_train) # 20.88 minutes
from dask_ml.model_selection import HyperbandSearchCV
dask_search = HyperbandSearchCV(
model, params, test_size=0.2, max_iter=max_iter, aggressiveness=4
)
from tune_sklearn import TuneSearchCV
ray_search = TuneSearchCV(
model, params, n_iter=n_params, max_iters=max_iter, early_stopping=True, **kwargs
)
dask_search.fit(X_train, y_train) # 2.93 minutes
ray_search.fit(X_train, y_train) # 2.49 minutes
Full example usage
from sklearn.linear_model import SGDClassifier
from scipy.stats import uniform, loguniform
from sklearn.datasets import make_classification
model = SGDClassifier()
params = {"alpha": loguniform(1e-5, 1e-3), "l1_ratio": uniform(0, 1)}
X, y = make_classification()
from sklearn.model_selection import RandomizedSearchCV
search = RandomizedSearchCV(model, params, ...)
search.fit(X, y)
from dask_ml.model_selection import HyperbandSearchCV
HyperbandSearchCV(model, params, ...)
search.fit(X, y, classes=[0, 1])
from tune_sklearn import TuneSearchCV
search = TuneSearchCV(model, params, ...)
search.fit(X, y, classes=[0, 1])
-
Their implementation of Hyperband in HpBandSter is included in Auto-PyTorch and BOAH. ↩
-
See Figures 4, 7 and 8 in “Hyperband: A Novel Bandit-Based Approach to Hyperparameter Optimization.” ↩
-
See Figure 1 of the BOHB paper and a paper from an augmented reality company. ↩
-
SMAC is described in “Sequential Model-Based Optimization forGeneral Algorithm Configuration,” and is available in AutoML. ↩
-
Spearmint is described in “Practical Bayesian Optimization of MachineLearning Algorithms,” and is available in HIPS/spearmint. ↩
-
TPE is described in Section 4 of “Algorithms for Hyperparameter Optimization,” and is available through Hyperopt. ↩
-
From Ray’s README.md: “If the estimator does not support
partial_fit, a warning will be shown saying early stopping cannot be done and it will simply run the cross-validation on Ray’s parallel back-end.” ↩ -
I choose to benchmark random searches instead of grid searches because random searches produce better results because grid searches require estimating how important each parameter is; for more detail see “Random Search for Hyperparameter Optimization” by Bergstra and Bengio. ↩
-
Despite a relevant implementation in dask-ml#527. ↩
-
Because priority is meaningless if there are an infinite number of workers. ↩
blog comments powered by Disqus