Load Large Image Data with Dask Array
By John Kirkham
Executive Summary
This post explores simple workflows to load large stacks of image data with Dask array.
In particular, we start with a directory full of TIFF files of images like the following:
$ $ ls raw/ | head
ex6-2_CamA_ch1_CAM1_stack0000_560nm_0000000msec_0001291795msecAbs_000x_000y_000z_0000t.tif
ex6-2_CamA_ch1_CAM1_stack0001_560nm_0043748msec_0001335543msecAbs_000x_000y_000z_0000t.tif
ex6-2_CamA_ch1_CAM1_stack0002_560nm_0087497msec_0001379292msecAbs_000x_000y_000z_0000t.tif
ex6-2_CamA_ch1_CAM1_stack0003_560nm_0131245msec_0001423040msecAbs_000x_000y_000z_0000t.tif
ex6-2_CamA_ch1_CAM1_stack0004_560nm_0174993msec_0001466788msecAbs_000x_000y_000z_0000t.tif
and show how to stitch these together into large lazy arrays using the dask-image library
>>> import dask_image
>>> x = dask_image.imread.imread('raw/*.tif')
or by writing your own Dask delayed image reader function.
|
Some day we’ll eventually be able to perform complex calculations on this dask array.
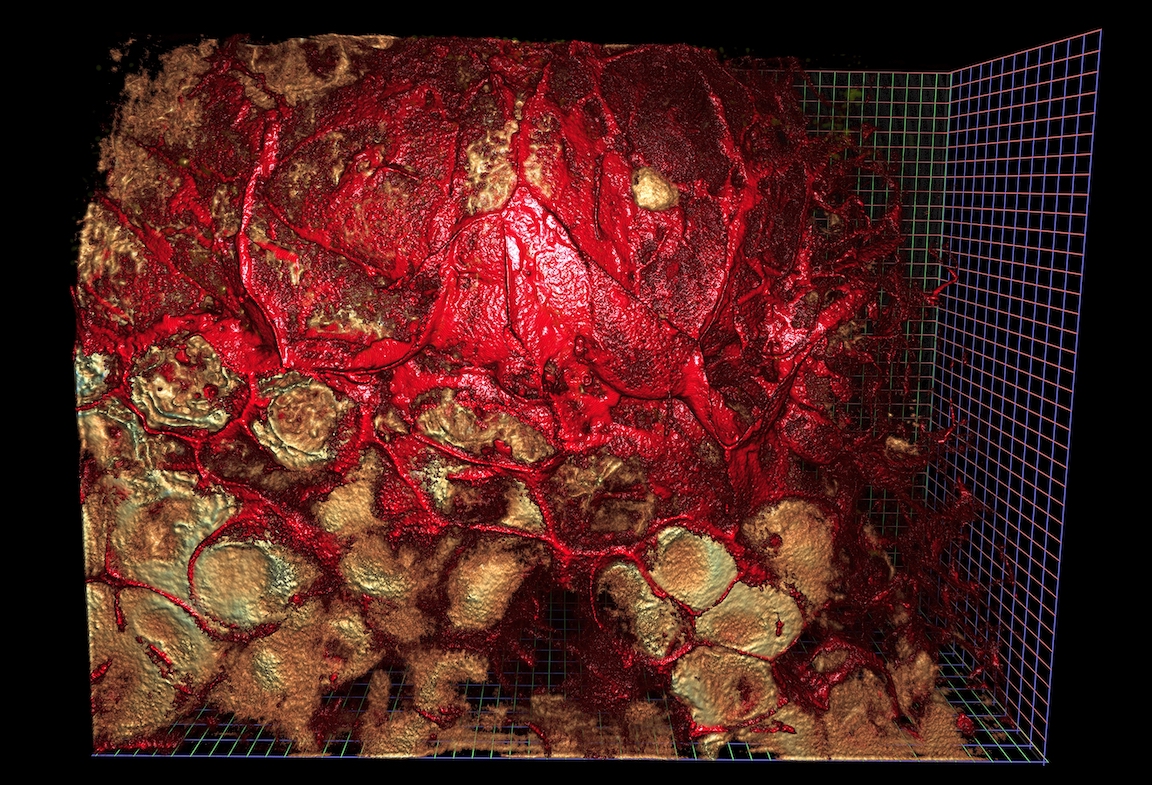
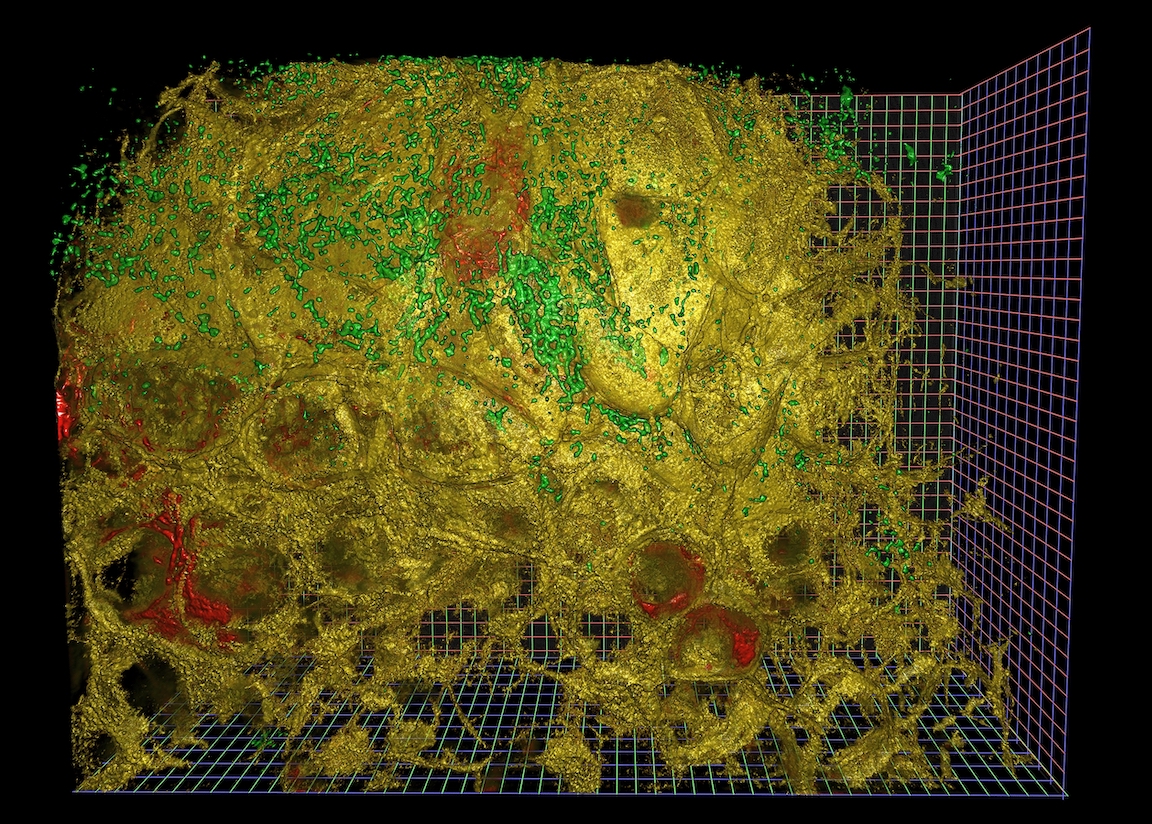
Disclaimer: we’re not going to produce rendered images like the above in this post. These were created with NVidia IndeX, a completely separate tool chain from what is being discussed here. This post covers the first step of image loading.
Series Overview
A common case in fields that acquire large amounts of imaging data is to write out smaller acquisitions into many small files. These files can tile a larger space, sub-sample from a larger time period, and may contain multiple channels. The acquisition techniques themselves are often state of the art and constantly pushing the envelope in term of how large a field of view can be acquired, at what resolution, and what quality.
Once acquired this data presents a number of challenges. Algorithms often designed and tested to work on very small pieces of this data need to be scaled up to work on the full dataset. It might not be clear at the outset what will actually work and so exploration still plays a very big part of the whole process.
Historically this analytical process has involved a lot of custom code. Often the analytical process is stitched together by a series of scripts possibly in several different languages that write various intermediate results to disk. Thanks to advances in modern tooling these process can be significantly improved. In this series of blogposts, we will outline ways for image scientists to leverage different tools to move towards a high level, friendly, cohesive, interactive analytical pipeline.
Post Overview
This post in particular focuses on loading and managing large stacks of image data in parallel from Python.
Loading large image data can be a complex and often unique problem. Different groups may choose to store this across many files on disk, a commodity or custom database solution, or they may opt to store it in the cloud. Not all datasets within the same group may be treated the same for a variety of reasons. In short, this means loading data is a hard and expensive problem.
Despite data being stored in many different ways, often groups want to reapply the same analytical pipeline to these datasets. However if the data pipeline is tightly coupled to a particular way of loading the data for later analytical steps, it may be very difficult if not impossible to reuse an existing pipeline. In other words, there is friction between the loading and analysis steps, which frustrates efforts to make things reusable.
Having a modular and general way to load data makes it easy to present data stored differently in a standard way. Further having a standard way to present data to analytical pipelines allows that part of the pipeline to focus on what it does best, analysis! In general, this should decouple these to components in a way that improves the experience of users involved in all parts of the pipeline.
We will use image data generously provided by Gokul Upadhyayula at the Advanced Bioimaging Center at UC Berkeley and discussed in this paper (preprint), though the workloads presented here should work for any kind of imaging data, or array data generally.
Load image data with Dask
Let’s start again with our image data from the top of the post:
$ $ ls /path/to/files/raw/ | head
ex6-2_CamA_ch1_CAM1_stack0000_560nm_0000000msec_0001291795msecAbs_000x_000y_000z_0000t.tif
ex6-2_CamA_ch1_CAM1_stack0001_560nm_0043748msec_0001335543msecAbs_000x_000y_000z_0000t.tif
ex6-2_CamA_ch1_CAM1_stack0002_560nm_0087497msec_0001379292msecAbs_000x_000y_000z_0000t.tif
ex6-2_CamA_ch1_CAM1_stack0003_560nm_0131245msec_0001423040msecAbs_000x_000y_000z_0000t.tif
ex6-2_CamA_ch1_CAM1_stack0004_560nm_0174993msec_0001466788msecAbs_000x_000y_000z_0000t.tif
Load a single sample image with Scikit-Image
To load a single image, we use Scikit-Image:
>>> import glob
>>> filenames = glob.glob("/path/to/files/raw/*.tif")
>>> len(filenames)
597
>>> import imageio
>>> sample = imageio.imread(filenames[0])
>>> sample.shape
(201, 1024, 768)
Each filename corresponds to some 3d chunk of a larger image. We can look at a few 2d slices of this single 3d chunk to get some context.
import matplotlib.pyplot as plt
import skimage.io
plt.figure(figsize=(10, 10))
skimage.io.imshow(sample[:, :, 0])
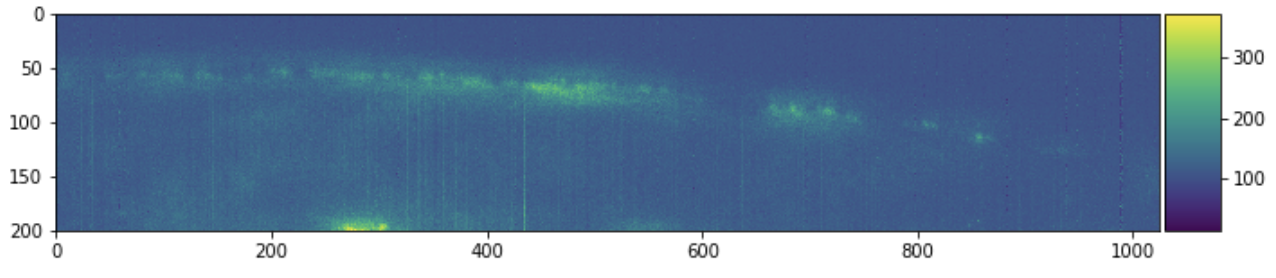
plt.figure(figsize=(10, 10))
skimage.io.imshow(sample[:, 0, :])
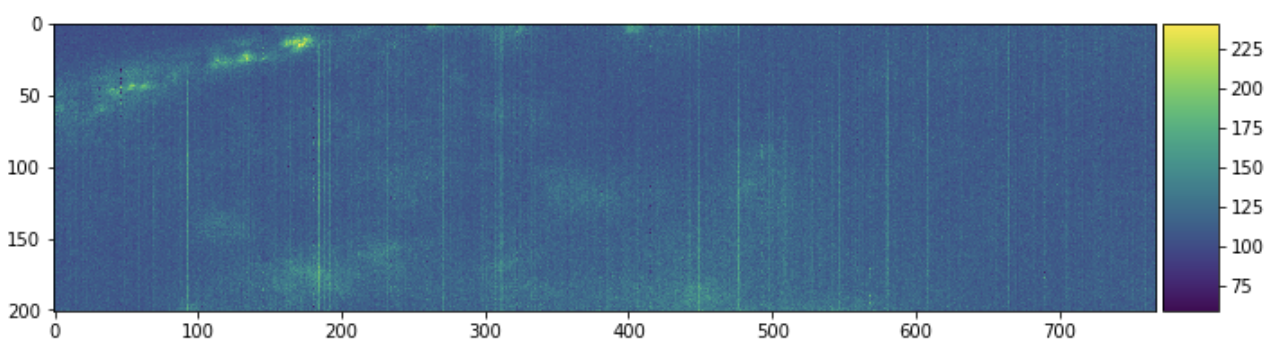
plt.figure(figsize=(10, 10))
skimage.io.imshow(sample[0, :, :])
Investigate Filename Structure
These are slices from only one chunk of a much larger aggregate image.
Our interest here is combining the pieces into a large image stack.
It is common to see a naming structure in the filenames. Each
filename then may indicate a channel, time step, and spatial location with the
<i> being some numeric values (possibly with units). Individual filenames may
have more or less information and may notate it differently than we have.
mydata_ch<i>_<j>t_<k>x_<l>y_<m>z.tif
In principle with NumPy we might allocate a giant array and then iteratively load images and place them into the giant array.
full_array = np.empty((..., ..., ..., ..., ...), dtype=sample.dtype)
for fn in filenames:
img = imageio.imread(fn)
index = get_location_from_filename(fn) # We need to write this function
full_array[index, :, :, :] = img
However if our data is large then we can’t load it all into memory at once like this into a single Numpy array, and instead we need to be a bit more clever to handle it efficiently. One approach here is to use Dask, which handles larger-than-memory workloads easily.
Lazily load images with Dask Array
Now we learn how to lazily load and stitch together image data with Dask array. We’ll start with simple examples first and then move onto the full example with this more complex dataset afterwards.
We can delay the imageio.imread calls with Dask
Delayed.
import dask
import dask.array as da
lazy_arrays = [dask.delayed(imageio.imread)(fn) for fn in filenames]
lazy_arrays = [da.from_delayed(x, shape=sample.shape, dtype=sample.dtype)
for x in lazy_arrays]
Note: here we’re assuming that all of the images have the same shape and dtype
as the sample file that we loaded above. This is not always the case. See the
dask_image note below in the Future Work section for an alternative.
We haven’t yet stitched these together. We have hundreds of single-chunk Dask arrays, each of which lazily loads a single 3d chunk of data from disk. Lets look at a single array.
>>> lazy_arrays[0]
|
This is a lazy 3-dimensional Dask array of a single 300MB chunk of data. That chunk is created by loading in a particular TIFF file. Normally Dask arrays are composed of many chunks. We can concatenate many of these single-chunked Dask arrays into a multi-chunked Dask array with functions like da.concatenate and da.stack.
Here we concatenate the first ten Dask arrays along a few axes, to get an
easier-to-understand picture of how this looks. Take a look both at how the
shape changes as we change the axis= parameter both in the table on the left
and the image on the right.
da.concatenate(lazy_arrays[:10], axis=0)
|
da.concatenate(lazy_arrays[:10], axis=1)
|
da.concatenate(lazy_arrays[:10], axis=2)
|
Or, if we wanted to make a new dimension, we would use da.stack. In this
case note that we’ve run out of easily visible dimensions, so you should take
note of the listed shape in the table input on the left more than the picture
on the right. Notice that we’ve stacked these 3d images into a 4d image.
da.stack(lazy_arrays[:10])
|
These are the common case situations, where you have a single axis along which you want to stitch images together.
Full example
This works fine for combining along a single axis. However if we need to combine across multiple we need to perform multiple concatenate steps. Fortunately there is a simpler option da.block, which can concatenate along multiple axes at once if you give it a nested list of dask arrays.
a = da.block([[laxy_array_00, lazy_array_01],
[lazy_array_10, lazy_array_11]])
We now do the following:
- Parse each filename to learn where it should live in the larger array
- See how many files are in each of our relevant dimensions
- Allocate a NumPy object-dtype array of the appropriate size, where each element of this array will hold a single-chunk Dask array
- Go through our filenames and insert the proper Dask array into the right position
- Call
da.blockon the result
This code is a bit complex, but shows what this looks like in a real-world setting
# Get various dimensions
fn_comp_sets = dict()
for fn in filenames:
for i, comp in enumerate(os.path.splitext(fn)[0].split("_")):
fn_comp_sets.setdefault(i, set())
fn_comp_sets[i].add(comp)
fn_comp_sets = list(map(sorted, fn_comp_sets.values()))
remap_comps = [
dict(map(reversed, enumerate(fn_comp_sets[2]))),
dict(map(reversed, enumerate(fn_comp_sets[4])))
]
# Create an empty object array to organize each chunk that loads a TIFF
a = np.empty(tuple(map(len, remap_comps)) + (1, 1, 1), dtype=object)
for fn, x in zip(filenames, lazy_arrays):
channel = int(fn[fn.index("_ch") + 3:].split("_")[0])
stack = int(fn[fn.index("_stack") + 6:].split("_")[0])
a[channel, stack, 0, 0, 0] = x
# Stitch together the many blocks into a single array
a = da.block(a.tolist())
|
That’s a 180 GB logical array, composed of around 600 chunks, each of size 300 MB. We can now do normal NumPy like computations on this array using Dask Array, but we’ll save that for a future post.
>>> # array computations would work fine, and would run in low memory
>>> # but we'll save actual computation for future posts
>>> a.sum().compute()
Save Data
To simplify data loading in the future, we store this in a large chunked array format like Zarr using the to_zarr method.
a.to_zarr("mydata.zarr")
We may add additional information about the image data as attributes. This both makes things simpler for future users (they can read the full dataset with a single line using da.from_zarr) and much more performant because Zarr is an analysis ready format that is efficiently encoded for computation.
Zarr uses the Blosc library for compression by default. For scientific imaging data, we can optionally pass compression options that provide a good compression ratio to speed tradeoff and optimize compression performance.
from numcodecs import Blosc
a.to_zarr("mydata.zarr", compressor=Blosc(cname='zstd', clevel=3, shuffle=Blosc.BITSHUFFLE))
Future Work
The workload above is generic and straightforward. It works well in simple cases and also extends well to more complex cases, providing you’re willing to write some for-loops and parsing code around your custom logic. It works on a single small-scale laptop as well as a large HPC or Cloud cluster. If you have a function that turns a filename into a NumPy array, you can generate large lazy Dask array using that function, Dask Delayed and Dask Array.
Dask Image
However, we can make things a bit easier for users if we specialize a bit. For example the Dask Image library has a parallel image reader function, which automates much of our work above in the simple case.
>>> import dask_image
>>> x = dask_image.imread.imread('raw/*.tif')
Similarly libraries like Xarray have readers for other file formats, like GeoTIFF.
As domains do more and more work like what we did above they tend to write down common patterns into domain-specific libraries, which then increases the accessibility and user base of these tools.
GPUs
If we have special hardware lying around like a few GPUs, we can move the data over to it and perform computations with a library like CuPy, which mimics NumPy very closely. Thus benefiting from the same operations listed above, but with the added performance of GPUs behind them.
import cupy as cp
a_gpu = a.map_blocks(cp.asarray)
Computation
Finally, in future blogposts we plan to talk about how to compute on our large Dask arrays using common image-processing workloads like overlapping stencil functions, segmentation and deconvolution, and integrating with other libraries like ITK.
blog comments powered by Disqus